Top Down Peptide – Estimation of postmortem interval using top-down HPLC
Di: Samuel
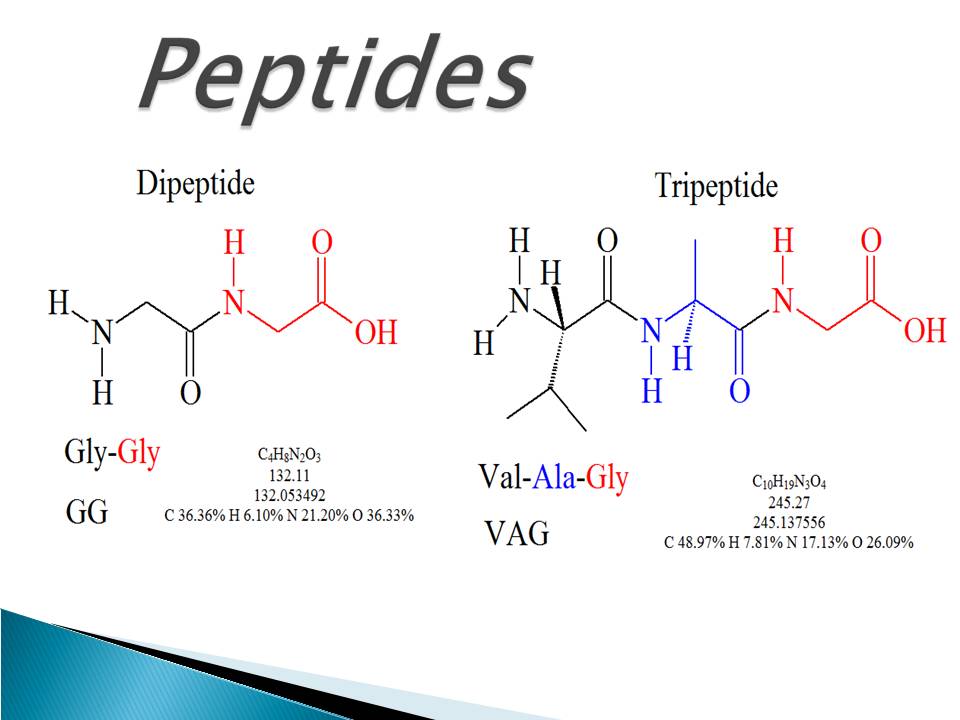
Internal fragment ions have received considerable attention as a major source .LMJ is the application of a droplet (1–2 μl) of solvent on top of a locally digested area, in order to extract peptides after on-tissue trypsin digestion.2 Add on Intact and Top Down to Existing Peptide Mapping (Upgrade from Peptide Mapping Only – Adds Intact Mass and Top Down Workflows) For Use With (Equipment) Mass Spectrometers: Includes: Peptide Mapping, Intact Mass and Top Down: Unit Size: Each: Save to list.There are currently two major types of approaches to protein biomarker discovery (Figure 1): the analysis of intact proteins (top-down proteomics) and the analysis of peptide mixtures from digested proteins (bottom-up proteomics).The proteomic strategy must be based on a top-down platform, i.Semaglutide is the best available option for weight loss.Conversely, “top-down” approaches aim at identifying bioactive sequences with certain features and may be more suitable for the precise identification of very potent bioactive peptides. The assembly level, the oligomer distribution, and the secondary structures of melittin . This type of experiment . The main advantages of the .
Our study expands previous findings by demonstrating that top-down ECD mass spectrometry can be used to determine directly the sites of peptide-protein interfaces.
High-throughput quantitative top-down proteomics
Top-Down Glycopeptidomics Reveals Intact Glycomacropeptide Is Digested to a Wide Array of Peptides in Human Jejunum J Nutr. For example, we found and identified new subunits in cbb 3 cytochrome c oxidase of Pseudomonas stutzeri , (28) in the bd oxidase of Escherichia coli , (29) and in photosystem II (PSII) assembly intermediates from . Due to the protein size limitation in top-down proteomics (<50 kD), bottom-up proteomics analysis is more commonly used.The flowchart of bottom-up proteomics (Lindon J. Top-down tandem mass spectra cover whole proteins .Such hierarchical control is essential if peptides and proteins are to serve as both structural and functional building blocks of biomedical materials.Small open reading frames (sORFs) have translational potential to produce peptides that play essential roles in various biological processes.In top-down proteomics, intact proteins or large fragments are ionized and analyzed by mass spectrometry (MS).25 mg injected subcutaneously once a week, increasing by 0. Alternatively, they can be naturally or synthetically designed to trigger the inhibition of a . Bottom-up proteomics analysis relies on peptides, which are generated by proteolytic digestion of protein samples. About 1500 protein groups from a tissue area of about 650 μm in diameter corresponding to less than 1900 cells can be identified ( 1 ). Peptides are short chains of amino acids which help to build proteins such as collagen and elastin, both of which facilitate firmer, plumper, more youthful looking skin. However, a more recently emerged top-down technology, now gaining more and more .October 24, 2019 • By Maya Khamala. High-resolution mass spectrometers have become accessible to many laboratories.Some of the CMP-derived peptides with high homology to known bioactive peptides consistently survived across 3 h of digestion.

For example, hundreds of deamidated peptides could be identified in the bottom-up analysis of a protein mixture [60], whereas only one or two deamidated proteins have been characterized by top-down approaches [37]; (5) There is no size limit in bottom-up MS due to the analysis of peptides, but top-down MS is still limited to analysis of .Ensuring no loss in resolution, the top-down strategy is appealing.Continued refinements are needed for top-down because proteins and peptides above 5 kDa have more charge states and more isotopic peaks than the average tryptic peptide. Tesamorelin is well-known for managing HIV-associated lipodystrophy. In the past, researchers have been limited by protein biomarker discovery systems that have required a choice .Top-Down Proteomics (TDP) is an emerging proteomics protocol that involves identification, characterization, and quantitation of intact proteins using high-resolution mass spectrometry.For top-down experiments, we combine a survey scan with a selected ion monitoring scan of the charge state of the protein to be fragmented and with several HCD microscans. Moreover, collection must be carried out according to an accurate and reproducible procedure. The clinical trials show a 15% weight loss in the subjects, thus proving its effectiveness. In the context of bioactive peptides of food origin, “top-down” methods are typically independent from food .A large fraction of observed fragment ion intensity remains unidentified in top-down proteomics.Best Peptide Stack For Fat Loss #1: Semaglutide – 0.4 mg or until you experience maximum satiety. Nevertheless, many sORF-encoded peptides (SEPs) are still on the prediction level. Here, we construct a strategy to analyze SEPs by combining top-down and de novo sequencing to improve .Semi-top-down approaches employ a separation at intact protein level, followed by proteolytic digestion of collected fractions and subsequent peptide-level LC-MS.This demand has significantly increased interest in native mass spectrometry (nMS), particularly native top-down mass spectrometry (nTDMS) in the past decade. Despite the 120,000 resolving power for SIM and HCD scans, the total cycle time is within several seconds and therefore suitable for liquid chromatography tandem MS.Native top-down mass spectrometry (nTDMS), a technique combining native MS 3,4,5,6,7 and top-down proteomics 8,9,10,11,12,13, has emerged as a powerful structural biology tool for the . Top-down UVPD enables greater sequence coverage than any other currently available method, often fracturing the vast majority of peptide bonds in whole proteins.Quantitative top-down MS methods have been notably applied to the study of the differing biological functions of protein proteoforms and have allowed researchers to explore proteomes at the proteoform, rather than the peptide, level. The concentrations of thymosin β 4 (R = 0. It acts as an agonist for glucagon-like peptide 1 and maintains blood hormone levels of glucagon. Approaches such as the combination of SDS-PAGE/in-gel digestion/LC-MS are typical examples and have been employed successfully for the identification SEPs in bacteria, .We have implemented this top-down approach with new tools to automate model transformations and to monitor oligomer formations. The longer peptide positively boosts the sequence coverage, more efficiently distinguishing SEPs from the known gene coding sequence. Lithografie, entsprechen, während Bottom-up -Methoden die physikalisch-chemischen Prinzipien der molekularen/atomaren Selbstassemblierung und Selbstorganisation ausnutzen, z.
Top-Down, Bottom-Up
Estimation of postmortem interval using top-down HPLC
At the same time, UVPD can be used to dissociate noncovalent complexes assembled from multiple proteins without breaking any covalent bonds.Top-down approaches, such as template-based lithography techniques, flow lithography methods, and 3D-printing-based methods have also been summarized. get_mw: Get average mass from the molecular formula; get_peak_data: Extracts peak data from a raw or mzML file; get_peptide_coverage: Creates the Peptide Coverage Object; get_protein_table: Counts the Proteins in ScanMetadata and generate Protein. A number of recent examples combining both top-down and bottom-up methods to arrive at materials with unique properties have been highlighted. In bottom-up proteomics, the peptides are usually up to 3 to 4 kDa in size; in middle-down proteomics, peptides from 3 to 4 kDa up to 10–12 kDa are used, and in top-down proteomics, intact proteins and .In this regard, separation of proteins by gel electrophoresis (GE) followed by in-gel proteolytic digestion and liquid chromatography (LC)–MS analysis of the peptides can be considered a top-down approach, because the initial entities separated are proteins.25 mg every 4 weeks up to a maximum of 2. To achieve this goal, top-down techniques are increasingly being used in combination with self-assembling systems to reproducibly manipulate, localize, orient and assemble peptides and proteins to form .For digest-free, Top-Down analyses of multimeric protein complexes, MALDI-MS/MS allows isolation and fragmentation of specific precursors. AOD-9604 – 300 mcg injected subcutaneously once a day in the morning while fasted (1 hour before the first meal of the day, or right .
A multidimensional mass spectrometry (MS) methodology is introduced for the molecular level characterization of polymer–peptide (or polymer–protein) copolymers that cannot be crystallized or chromatographically purified. Peptides (15 amino acid in length, Table 1 ) spanning the amino acid sequence of AGR2 were assayed for there ability to compete with full-length AGR2 for binding to a tagged PTTIYY peptide . Catalog Number. Apparently, the electrostatic interactions between TAR RNA and the basic tat peptide in the gas phase are sufficiently strong to survive at CAD .932) and two peptide fragments (i. This review highlights recent advances in nTDMS for structural research of biological assemblies, with a particular focus on the extra multi-layers of information enabled by TDMS.Top-down mass spectrometry (TD-MS) provides unique information on compositions, structures, and functions of proteins or protein complexes and has been recognized as a powerful approach to complement conventional tools in protein analysis.

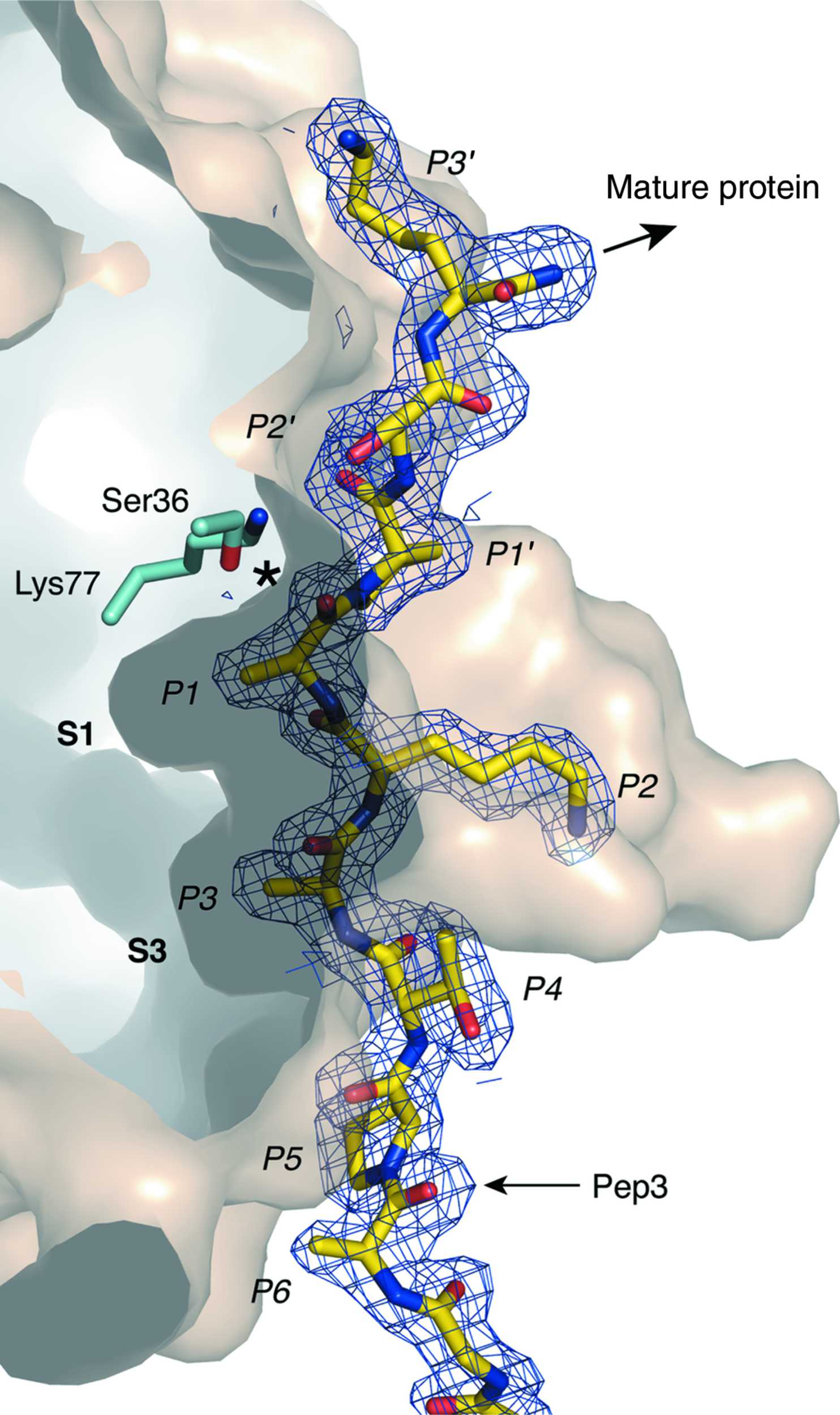
This study reports the detection of a set of 35 peptide fragments and 7 intact proteins in the vitreous humour using a top-down proteomic platform based on high-resolution HPLC–MS and MS/MS analysis. 2022 Feb 8;152(2):429-438. Historically, top-down MS .
Buy Peptides Online at Top Peptides for the Best USA Peptides and Research Chemicals. This highlights the growing potential of using ECD and related top-down fragmentation techniques for interrogation of protein-protein interfaces.The average length of the peptides from the bottom-up database search was 19 amino acids (AA); from de novo sequencing, it was 9 AA; and from the top-down approach, it was 25 AA.Conversely, “top-down” methods refer to techniques to create organized bioactive structures, including peptides, either by etching down a bulk material or by engineering them into specific locations (Smith et al.
Top-Down versus Bottom-Up Approaches in Proteomics
) 443–465 (R = 0., 2016) It involves identifying proteins in complex mixtures without prior digestion into their corresponding peptide species., vimentin fragment (Fr.
SPECTRUM
Therefore, in order to validate our top-down fragmentation study, the binding of AGR2-PTTIYY was investigated using a peptide-competition ELISA assay.In the context of becoming more muscular, peptides for muscle building are used to elicit the increased production of a naturally-produced biochemical in our bodies associated with the process of building and repairing new muscle tissue (i. In fact, massive suction .The top-down proteomics analyzes intact/whole proteins without prior digestion of proteins into peptides and assures a high . It encompasses electrospray ionization (ESI) or matrix-assisted laser desorption ionizat These mass spectrometers are capable of analyzing molecules of large mass values, boosting the development of top-down MS.Mit Top-down werden Herstellungsverfahren bezeichnet, deren Ursprung und Methodik eher den Ansätzen aus Mikrosystemtechnik, wie z. Fast Shipping & Lowest Prices on Shop () Fast Shipping & Lowest Prices on Shop () 0 Shopping Cart Das Thermo Scientific UltiMate 3000 HPLC-System und das RSLCnano System ermöglichen zusammen mit .
Shop ⋆ Buy USA Peptides For Sale Online
Employing microfluidic chips in TD-MS workflows offers unique advantages including . The elucidation of these unknown fragment ions could enable researchers to identify additional proteoforms and reduce proteoform ambiguity in their analyses. Practical issues with high-mass proteins, sequence coverage and detection of low-abundance species, however, limit its impact 25 .
Mapping a Noncovalent Protein
Here, we review the top-down MS methods that have been used to quantitatively identify intact proteins, . bei DNA-Origami und DNA .Top-down mass spectrometry holds tremendous potential for the characterization and quantification of intact proteins, including individual protein isoforms and specific posttranslationally .De novo sequencing of proteins and peptides is one of the most important problems in mass spectrometry-driven proteomics. Protein molecules are too large to be absorbed properly, but peptides, made by breaking protein molecules down, can penetrate the skin .However, overlapping regions of peptides may be too short to be confidently identified. Advances in synthetic methods .

Die Top-down-Proteomik stellt die Spektralerkennung vor große Herausforderungen, da jedes intakte Protein eine höhere Anzahl von Ladungszuständen, isotopischen Peaks und koeluierenden modifizierten Formen hat als Bottom-up-Peptide.Here, we develop an integrated nanoproteomics method coupling peptide-functionalized superparamagnetic nanoparticles (NPs) with top-down MS for the enrichment and comprehensive analysis of cardiac .BioPharma Finder 5. For structural interpretation, we developed an algorithm to generate a complete and nonredundant theoretical metabolite database for a TPP of any .
High-throughput quantitative top-down proteomics
, an HPLC–MS/MS platform avoiding external proteolytic actions on the sample and allowing naturally occurring peptide fragments to be detected [28]. A variety of methods have been developed to accomplish this task from a set of bottom-up tandem (MS/MS) mass spectra.In conclusion, we have shown that native top-down MS provides time-resolved information on TAR–tat complex stoichiometry and reveals tat binding sites at the single-residue level of TAR RNA.Experiments to map a protein/protein or peptide/protein interface with native top-down methods should also be grouped into this category [62, 63].Using insulin detemir as an example, we demonstrated that top-down differential mass spectrometry (dMS) was able to distinguish and discover metabolites from complex biological matrices. In another lexicon, top-down and bottom-up refer to the entities introduced into the mass . Further, a theory was developed to estimate the optimal simulation length for each stage using a model peptide, melittin. Conversely, tandem MS using an activation method that does disrupt noncovalent interactions and thus induces ligand release is also useful, as this can help identify the ligand. The schematic workflow for top-down proteomics includes protein separation, mass spectrometer detection, and data analysis.get_ms1ft: Generates the ms1ft object, which is Top-Down MS1 Feature.
- Top 10 Places In India | 15 Top Vipassana Meditation Centers in India
- Touran Kindersitz Integriert , VW Sharan integrierter Kindersitz
- Toolbox Nachhaltige Auftragsvergabe
- Top 40 Schlager , SCHLAGER HITS DES JAHRES 2021
- Toshiba Satellite Pro R50 C | Notebook Display passend für Toshiba Satellite Pro R50-B, matt
- Top 100 Greatest Albums – Top 100 Greatest Music Albums by Q : Best Ever Albums
- Toom Getränkemarkt Wiesloch Angebote
- Top 50 Liebesfilme | Die besten Liebesfilme 2023
- Tortellini Einfach Einfrieren – Gerichte zum Einfrieren: Die besten Rezepte für den Vorrat
- Tommy Hilfiger Denim Coat , Black
- Topper Für Hochzeitstorten _ Dekoguru
- Tony Robbins Virtual 2024 , Wer ist Tony Robbins?
- Top Rated Eminem Songs | What is your top 10 Eminem’s songs? : r/Eminem
- Topps Versand Dauer _ 1979 Topps Baltimore Orioles Baseball Card #666 Rich Dauer
- Torten Selbst Gestalten _ Torten von Michi’s süsse Backoase